The structure and phylogenetic affiliation of the Toc and Tic subunits in plastids of Plantae reflects their origin through primary endosymbiosis.
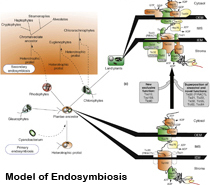
a| A cyanobacterium was captured by a mitochondriate eukaryote (i.e. a heterotrophic protist) and transformed into the plastid. This primary endosymbiotic event resulted in the ancestor of the algal lineages Chlorophyta, Rhodophyta and Glaucophyta. Land plants are derived from chlorophyte algae. Secondary endosymbiotic events resulted in the chromalveolate, euglenophyte and chlorarachniophyte algae when a heterotrophic protist engulfed an existing alga with a "primary" plastid.
b| The Toc and Tic translocons of higher plants in the process of importing a pre−protein from the cytosol are shown from the perspective of the chloroplast envelope membranes. The unfolded cytosolic proteins containing a TP (N−terminal segment) are initially recognized by the receptors Toc34 and Toc159 and then imported across the outer envelope membrane (OEM) and inner envelope membrane (IEM) of plastids through the pore components Toc75 and Tic110. The translocation process is energy dependent (ATP hydrolysis) and is driven by heat shock−type molecular chaperones (Hsp70 and Hsp93) acting at the cytosolic intermembrane space (IMS) and stromal sides of the organelle. A stromal processing peptidase (SPP) proteolytically removes the TP from post−translocated polypeptides. Several subunits are functionally engaged in the Toc and Tic translocons.
c| The proposed primitive state of the translocon in the Plantae ancestor was simple and composed of the Toc34, Toc75 and Tic110 subunits and the Hsp70 and Hsp93 chaperones. The hypothetical inclusion of the Tic20 and Tic21 subunits in the primitive translocon is still uncertain and depends on direct experimental proof of their participation as channel components of the Tic translocon. This transition involved the addition of subunits to fulfil an exclusive new function in protein import (Toc12, Toc159 and Tic40) or subunits that were functionally recruited to the translocon but maintained ancestral properties. We tentatively classify Toc64 in the latter group because it shows sequence conservation with plant amidases. We raise the question of whether the protein import−related anion channel (PIRAC) corresponds to Tic20.
Molecular factors are either encoded in the nucleus or in the alphaproteobacterial or cyanobacterial chromosome.
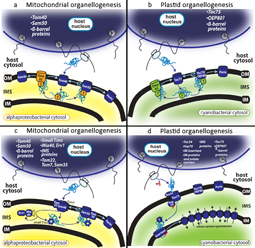
a,b| Genes encoding Tom40, Sam50, Toc75 and possibly OeP80 were relocated to the nuclear genome of the respective eukaryotic hosts and the encoded mRNA translated by cytosolic ribosomes. The resulting proteins were directed across the Tom40 pore in the alphaproteobacterial outer membrane (OM) or the Toc75 channel in the cyanobacterial OM, then protected by bacterial periplasmic chaperones in the intermembrane space (iMS) and assembled in the OM by endogenous Omp85. Accumulated OM pools of Sam50 and Toc75 (or OeP80) increasingly assumed control over the assembly of nuclear−encoded proteins, enabling the establishment in the nucleus of new genes for organelle−destined β−barrel proteins.
c,d| Novel nuclear−encoded factors gained access to the iMS, replaced the endogenous components and supported the biogenesis of β−barrels by Sam50 in the alphaproteobacterial OM and Toc75 (or OeP80) in the cyanobacterial OM. erv1 and Mia40 were introduced during mitochondrial evolution as a disulphide relay system for newly established iMS proteins, including the ancestor of the small Tim chaperones and Mia40−erv1. At this stage, the subunits Tom22 and Sam35 could have been added as receptors for β−barrel proteins and Tom7 as a negative regulator of Tom40 biogenesis.
Molecular factors are either encoded in the nucleus or in the alphaproteobacterial or cyanobacterial chromosome.
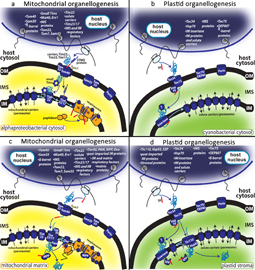
a| The Tim22 insertase was introduced in the alphaproteobacterial inner membrane (iM) to catalyse integration of the mitochondrial carriers and its own insertion. Small Tim proteins bridged the transit of sorting substrates from Tom40 to the Tim22 insertase. Tim23 and Tim17 were substrates of Tim22 inserted in the iM to constitute another protein pore. This pore evolved to integrate STMD proteins containing presequences into the iM as respiratory factors to control the biogenesis of the alphaproteobacterial respiratory Complex i, Complex iii and Complex iv (the respirasome). An alphaproteobacterial cytosolic peptidase was recruited to the respirasome Complex iii to cleave the presequences of Tim23 substrates. Other soluble respiratory factors could potentially have been attached to the sector of the respirasome exposed to the iMS.
b| A putative nuclear−encoded insertase evolved to integrate nuclear−encoded solute carriers in the cyanobacterial iM. Its function was supported by hypothetical nuclear−encoded components (chaperones) that were previously established in the iMS. At this stage the transit peptide TPs could have evolved either to facilitate the translocation of substrates across the outer membrane (OM) by binding an iMS Hsp70 motor, or to help the integration of solute carriers by the iM insertase. Toc34 was established as a receptor for TP−containing proteins.
c| The putative ancestral protein−sorting system in mitochondria. The addition of the presequence translocase−associated motor (PAM) module to the Tim23 insertase allowed the translocation of proteins into the mitochondrial matrix. Among them were new respiratory factors acting in the respirasome sector exposed to the mitochondrial matrix. Matrix processing peptidase (MPP) was derived from the alphaproteobacterial peptidase when the corresponding gene was established in the nucleus. Pools of MPP became associated with the repirasome complex iii. Oxa was established as a mitoribosome recruiting factor to promote the cotranslational insertion of respirasome subunits encoded in the mitochondrial genome. In addition, Oxa provided a pathway for post−import insertion in the iM of nuclear−encoded proteins.
d| The putative ancestral protein−sorting system in plastids. The Tic110 translocase evolved in connection with the Hsp93 chaperone motor, making possible the translocation of proteins into the plastid stroma. Among them was the nuclear−encoded stromal processing peptidase (SPP). The Tic complex might have provided a pathway for post−import insertion of iM proteins such as solute carriers and Tic110.