Project Description
Endosymbiosis, plastid evolution and the origin of Plantae
Early in eukaryotic evolution, a heterotrophic unicellular eukaryote engulfed a cyanobacterium and retained it in its cytoplasm, converting it into the plastid.
This event is referred to as "primary endosymbiosis" and the photosynthetic eukaryote putatively gave rise to the common ancestor of the "supergroup" Plantae: the Glaucophyta, the Rhodophyta and the Viridiplantae (green algae and land plants).
The number of independent primary endosymbioses has profound implications for our understanding of the evolution of eukaryotes. If the Plantae members had independent primary endosymbioses (i.e., as many as three times), that would suggest that establishing the combination of a eukaryotic and a prokaryotic cell is relatively easy, in evolutionary terms, and the resulting chimera inevitably converged on a similar morphology and physiology for each capture.
If it occurred only once, then all extant autotrophic eukaryotes trace their ancestry to this remarkable "accident" of evolution that gave rise to a vast assemblage of organisms that came to dominate the planet and become the driving force behind its climate, geochemistry, and ecology.
Whereas it has not yet been definitively proven, the majority of phylogenetic and genetic evidence suggests that this primary endosymbiosis occurred a single time in the common ancestor of the Plantae and all plastids in eukaryotes trace their ancestry to this singular event.
Gene transfer and footprints of endosymbiosis
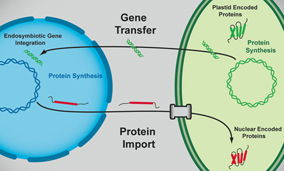 A critical step in plastid establishment is the "gene transfer" from the endosymbiont to the host nuclear genome. It is clear that if two organisms live in close association, but each contains its own complete complement of genes, these cells can live apart, if necessary. When a gene is required for the viability of the endosymbiont, it is transferred to the nucleus of the host, the two associated organisms can no longer live separately. If these transferred genes code for proteins that function inside the endosymbiont, then a system for importing proteins is synthesized in the cytoplasm must also be developed.
A critical step in plastid establishment is the "gene transfer" from the endosymbiont to the host nuclear genome. It is clear that if two organisms live in close association, but each contains its own complete complement of genes, these cells can live apart, if necessary. When a gene is required for the viability of the endosymbiont, it is transferred to the nucleus of the host, the two associated organisms can no longer live separately. If these transferred genes code for proteins that function inside the endosymbiont, then a system for importing proteins is synthesized in the cytoplasm must also be developed.
Most plastids contain a single, circular chromosome of about 200 Kb and encode around 100−120 genes. A free−living cyanobacterium typically has a genome of about 4000−5000 Kb. As consequence of endosymbiotic gene transfer and gene losses, plastid chromosomes are the outcome of an extreme genome reduction process. The genes that remain in the plastid are primarily involved in photosynthesis or transcription and translation of plastid genes.
However, most genes that needed to maintain the plastid are encoded in the nucleus. Plastid−encoded genes are presumably under selection for transfer to the nucleus to avoid the accumulation of deleterious mutations that occur in non−recombining genomes. The completion of several cyanobacterial and eukaryotic genomes has allowed us to understand the impact of this endosymbiosis more completely on the genome of eukaryotes. Analyses of unicellular algal genomic data, including Expressed sequence tag (EST) analysis of Cyanophora paradoxa, indicate that between 600−800 cyanobacterial genes have been transferred and retained in the nucleus during Plantae evolution and the vast majority encode plastid−targeted proteins.
These results are in contrast with the findings in Arabidopsis of as much as 4500 cyanobacterial genes may be present in their nuclear genome and ca. the 50% of them are predicted as non−plastid targeted proteins.
Why sequence the Cyanophora paradoxa genome?
Glaucophytes have been largely considered living fossils among photosynthetic eukaryotes, even affectionately called the "coelacanth" of the algal world (John M. Archibald). This venerable consideration reflects the retention of unique cyanobacterial characters in their plastids (cyanelles), such as the presence of remnants of the peptidoglycan cell wall and carboxysomes which are considered "primitive" traits putatively present in the plastid of the Plantae common ancestor.
Consistent with this evidence, our recent phylogenetic work suggests that glaucophytes diverged from the Plantae stem before the split of green algae and red algae. Historically, most biological, biochemical and molecular studies of glaucophytes have focused on Cyanophora paradoxa. And in recent years, our group and others have generated several thousands of Expressed sequence tags. In summary, the current knowledge and available data point to Cyanophora paradoxa as being an ideal choice for genome sequencing to study the evolution of Plantae.
 A critical step in plastid establishment is the "gene transfer" from the endosymbiont to the host nuclear genome. It is clear that if two organisms live in close association, but each contains its own complete complement of genes, these cells can live apart, if necessary. When a gene is required for the viability of the endosymbiont, it is transferred to the nucleus of the host, the two associated organisms can no longer live separately. If these transferred genes code for proteins that function inside the endosymbiont, then a system for importing proteins is synthesized in the cytoplasm must also be developed.
A critical step in plastid establishment is the "gene transfer" from the endosymbiont to the host nuclear genome. It is clear that if two organisms live in close association, but each contains its own complete complement of genes, these cells can live apart, if necessary. When a gene is required for the viability of the endosymbiont, it is transferred to the nucleus of the host, the two associated organisms can no longer live separately. If these transferred genes code for proteins that function inside the endosymbiont, then a system for importing proteins is synthesized in the cytoplasm must also be developed.